Overview
This addition will allow users to see the history of directives made to a column of data.
Goals
User should be able to see lineage information, ie. directives, for columns
Storing lineage information should have minimal/no impact to the wrangler application
User Stories
As a user, I should be able to see the directives applied to a column of data.
As a user, I should be able to see the directives applied to a column of data over any period of time.
- As a user, I should be able to see how any column got to its current state as well as other columns that were impacted by it
As a user, I should be able to add tags and properties to specific columns of data (stretch)
Design
Save directives for each column in AST format after parsing of directives along with input and output schemas.
prepareRun() api to send information to platform.
***release 4.4 onwards implemenations can be found in branches feature/field-level-lineage-api and feature/field-level-lineage-graph***
Unmarshal and connect into graph and store in HBase.
Access to lineage should only be available through the platform
Approach
- Each directive implements `public MutationDefinition lineage()`
- prepareRun() function in Wrangler.java creates FieldLevelLineage instance and sends it to CDAP
- CDAP receives all instances of FieldLevelLineage from all transform stages and takes 4 variables to build graph:
- ProgramRunId
- PipelinePhase
- Mapping of stage name to FieldLevelLineage for all transform stages
- Mapping of stage name to Dataset name for sources and sinks
- Graph is generated and stored in HBase in a way that is easily accessible to UI
API changes
New Programmatic APIs:
TransformStep:
TransformStep is a Java interface that represents a modification done to a dataset by a transform stage.
/**
* <p>
* A TransformStep represents a modification done by a Transform.
* </p>
*/
public interface TransformStep {
/**
* @return the name of this modification
*/
String getName();
/**
* @return additional information about this modification
*/
@Nullable
String getInformation();
}
FieldLevelLineage:
FieldLevelLineage is a Java interface that contains all the necessary field-level lineage information to be sent to the CDAP platform, for transform stages.
import java.util.List;
import java.util.Map;
/**
* <p>FieldLevelLineage is a DataType for computing lineage for each field in a dataset.
* An instance of this type can be sent to be sent to platform through API for transform stages.</p>
*/
public interface FieldLevelLineage {
/**
* <p>A BranchingTransformStepNode represents a linking between a {@link TransformStep} and a field of data.
* It contains data about how this TransformStep affected this field.</p>
*/
interface BranchingTransformStepNode {
/**
* @return the index of the TransformStep in {@link #getSteps()}
*/
int getTransformStepNumber();
/**
* @return true if this TransformStep does not add this field
*/
boolean continueBackward(); // continue down
/**
* @return true if this TransformStep does not drop this field
*/
boolean continueForward(); // continue up
/**
* This map should contain every other field that was impacted by this field in this TransformStep
* @return A map from field name to the index of the next TransformStep using {@link #getLineage()}
* Usage: getLineage().get(field).get(index).
*/
Map<String, Integer> getImpactedBranches(); // getUpBranches
/**
* This map should contain every other field that impacted this field with this TransformStep
* @return A map from field name to the index of the previous transformation step in {@link #getLineage()}
* Usage: getLineage().get(field).get(index).
*/
Map<String, Integer> getImpactingBranches(); // getDownBranches
}
/**
* @return a list of all TransformSteps executed by this Transform in order.
*/
List<TransformStep> getSteps();
/**
* @return a mapping of field names to a list of {@link BranchingTransformStepNode} in reverse order.
*/
Map<String, List<BranchingTransformStepNode>> getLineage();
}
WranglerFieldLevelLineage:
Instance should be initialized per wrangler node passing in a list of start and end columns (input and output schema).
build() takes a List<MutationDefinition> and stores all the necessary information into lineages.
Stores lineage for each column in lineage instance variable which is a map to ASTs.
/**
* WranglerFieldLevelLineage is a data type for computing lineage for all columns in Wrangler.
* An instance of this type to be sent to platform through prepareRun API.
*/
public final class WranglerFieldLevelLineage implements FieldLevelLineage {
/**
* A WranglerBranchingStepNode is a data type for linking columns to directives as well as other columns.
*/
public final class WranglerBranchingStepNode implements BranchingTransformStepNode {
private final int stepNumber;
private boolean continueUp;
private boolean continueDown;
private final Map<String, Integer> upBranches;
private final Map<String, Integer> downBranches;
private WranglerBranchingStepNode(int stepNumber, boolean continueUp, boolean continueDown) {
this.stepNumber = stepNumber;
this.continueUp = continueUp;
this.continueDown = continueDown;
this.upBranches = new HashMap<>();
this.downBranches = new HashMap<>();
}
// Internal functions
@Override
public int getTransformStepNumber() {
return stepNumber;
}
@Override
public boolean continueBackward() {
return continueUp;
}
@Override
public boolean continueForward() {
return continueDown;
}
@Override
public Map<String, Integer> getImpactedBranches() {
return Collections.unmodifiableMap(downBranches);
}
@Override
public Map<String, Integer> getImpactingBranches() {
return Collections.unmodifiableMap(upBranches);
}
@Override
public String toString() {
return "(StepNumber: " + stepNumber + ", Continue Backward: " + continueUp + ", " + upBranches +
", Continue Forward: " + continueDown + ", " + downBranches + ")";
}
}
private final List<TransformStep> steps;
private final Map<String, List<BranchingTransformStepNode>> lineage;
// Internal functions
// Shows lineage in tree format
void prettyPrint(String column, boolean forward) {...}
@Override
public List<TransformStep> getSteps() {
return Collections.unmodifiableList(this.steps);
}
@Override
public Map<String, List<BranchingTransformStepNode>> getLineage() {
return Collections.unmodifiableMap(this.lineage);
}
@Override
public String toString() {
return "Column Directives: " + lineage + "\n" + "Steps: " + steps;
}
/**
* Builder class for {@link WranglerFieldLevelLineage}.
*/
public static class Builder {
// Internal functions
public Builder(List<String> startColumns, List<String> finalColumns) {...}
public WranglerFieldLevelLineage build(List<MutationDefinition> parseTree) throws LineageGenerationException {...}
}
}
parseTree should contain all columns affected per directive.
Labels:
- All columns should be labeled one of: {Read, Drop, Modify, Add, Rename}
- Read: column's name or values are read. Including reading values and modifying. ie. "filter-rows-on..."
- Drop: column is dropped
- Add: column is added
- Rename: column's name is replaced with another name
- Modify: column's values altered and doesn't fit in any of the other categories, ie. "lowercase"
For Read, Drop, Modify, and Add the column and associated label should be something like -> Column: Name, Label: add.
For Rename the column and associated label should be something like -> Column: body_5 DOB, Label: rename. // Basically some way of having both names, currently using a space. Old/new for rename. Swap example: A B, Label: rename and B A, Label: rename.
**Assumption (can be changed): In ParseTree all columns should be in order of impact. ie. If directive is "copy A A_copy". "A, Label: read" should be before "A_copy, Label: add".
Algorithm visual: Example wrangler application --> forward lineage --> backward lineage
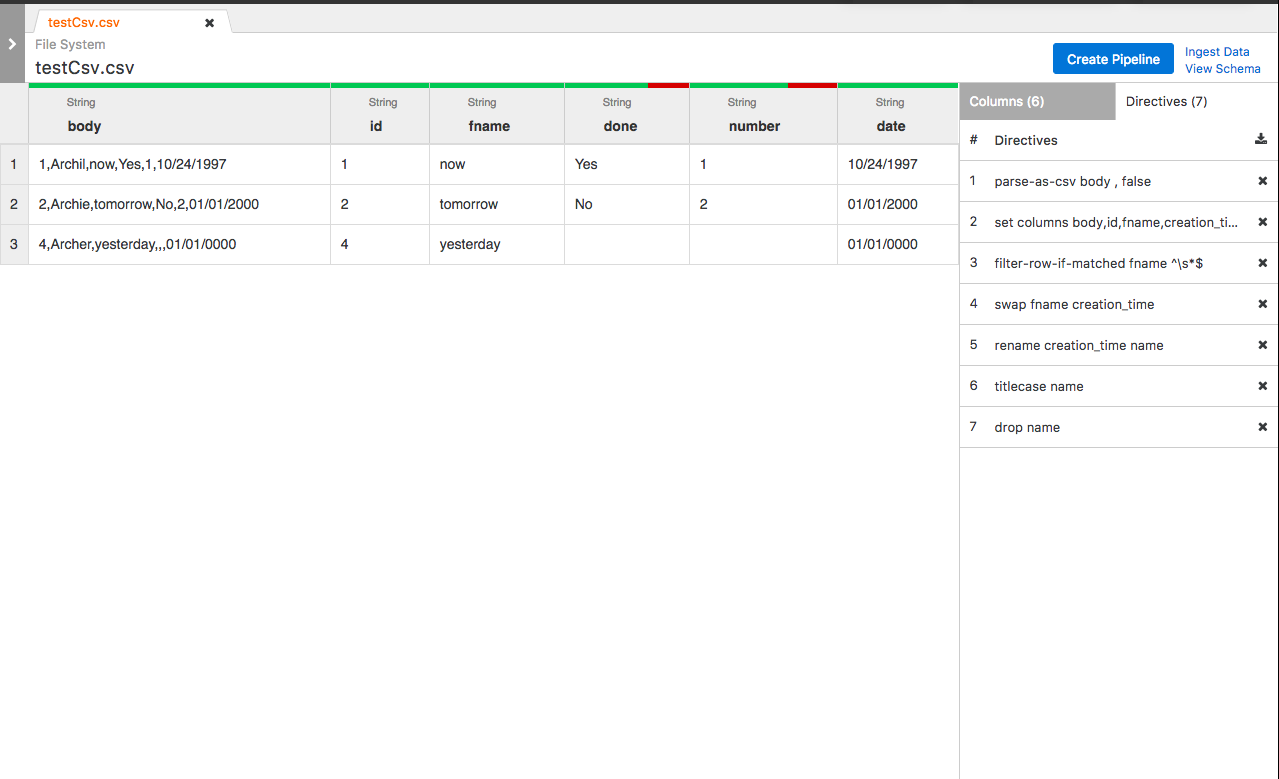
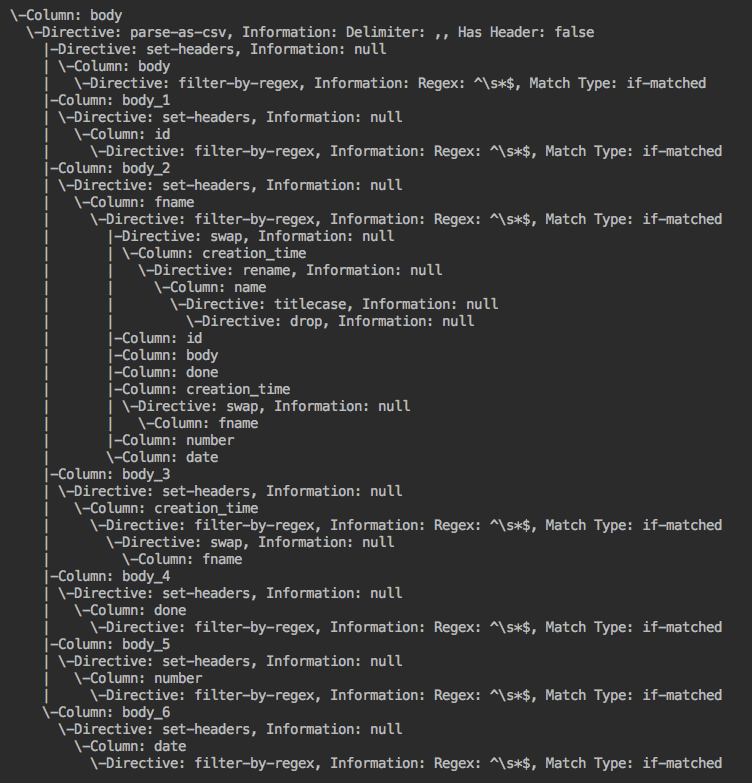
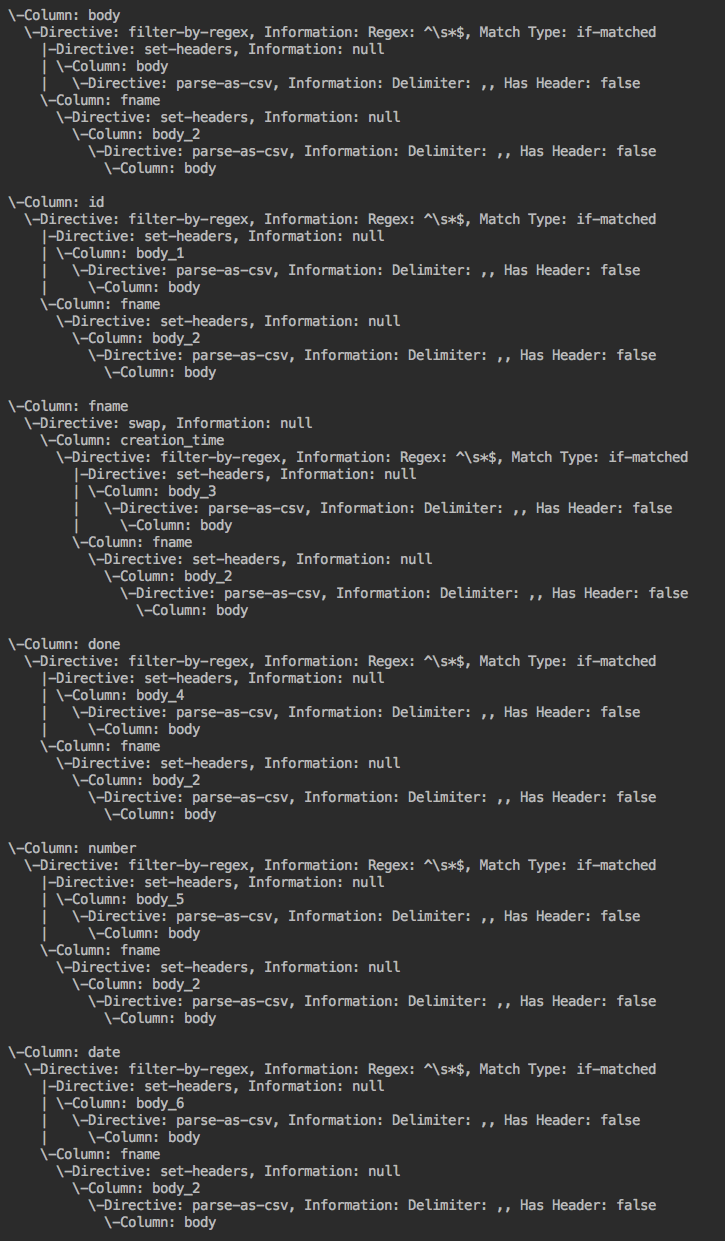
FieldLevelLineageStorageNode (IN BRANCH feature/field-level-lineage-storage):
FieldLevelLineageStorageNode is a Java interface that represents an element being stored in HBase.
These nodes are created and connected in an AST format by FieldLevelLineageStorageGraph.
/**
* <p>A FieldLevelLineageStorageNode represents a node of field-level lineage information.</p>
*/
public interface FieldLevelLineageStorageNode {
/**
* @return the ID of the pipeline
*/
ProgramRunId getPipeline();
/**
* @return the name of the stage
*/
String getStage();
/**
* @return the name of the field
*/
String getField();
}
FieldLevelLineageStorageGraph (IN BRANCH feature/field-level-lineage-storage):
FieldLevelLineageStorageGraph is a Java class that transforms many instances of FieldLevelLineage into a graph of FieldLevelLineageStorageNodes for storage.
One instance of this type should be created per pipeline. Created from many instances of FieldLevelLineage, some graph representation of the pipeline, a mapping between reference names and stage names for sources/sinks, and ProgramRunId.
import com.google.common.collect.ArrayListMultimap;
import com.google.common.collect.HashBasedTable;
import com.google.common.collect.ListMultimap;
import com.google.common.collect.Table;
import java.util.HashMap;
import java.util.List;
import java.util.Map;
public final class FieldLevelLineageStorageGraph {
private final ProgramRunId pipelineId;
private final PipelinePhase pipeline; // Or another type of graph of the pipeline
private final Map<String, FieldLevelLineage> stages;
private final Map<String, String> stageToDataSet;
private final Map<FieldLevelLineageStorageNode, FieldLevelLineageStorageNode> nodeRetriever;
private final Table<String, String, DataSetFieldNode> history; // Storage
private final Map<FieldStepNode, TransformStep> stepInformation; // Storage
private final ListMultimap<FieldLevelLineageStorageNode, FieldLevelLineageStorageNode> pastEdges; // Storage
private final ListMultimap<FieldLevelLineageStorageNode, FieldLevelLineageStorageNode> futureEdges; // Storage
public FieldLevelLineageStorageGraph(ProgramRunId PipelineId, PipelinePhase pipeline,
Map<String, FieldLevelLineage> stages, Map<String, String> stageToDataSet) {
this.pipelineId = PipelineId;
this.pipeline = pipeline;
this.stages = stages;
this.stageToDataSet = stageToDataSet;
this.nodeRetriever = new HashMap<>();
this.history = HashBasedTable.create();
this.stepInformation = new HashMap<>();
this.pastEdges = ArrayListMultimap.create();
this.futureEdges = ArrayListMultimap.create();
}
// helpers for make()
private void make() {...} // makes the graph
}
Visual:

Publishing Lineage Information:
Will be done through new prepareRun() function in the Transform class and new recordLineage(FieldLevelLineage f) function in StageSubmitter interface. Each transform plugin stage will have option to publish field-level lineage.
Wrangler Example:
...
@Override
public void prepareRun(StageSubmitter<TransformContext> context) throws Exception {
if (context.getContext().getArguments().has("enable.wrangler.field.lineage")) {
List<String> startColumns, endColumns;
Schema inputSchema, outputSchema;
inputSchema = context.getContext().getInputSchema();
outputSchema = context.getContext().getOutputSchema();
if (inputSchema == null || inputSchema.getFields() == null) {
startColumns = new ArrayList<>(0);
} else {
startColumns = new ArrayList<>(inputSchema.getFields().size());
for (Schema.Field field : inputSchema.getFields()) {
startColumns.add(field.getName());
}
}
if (outputSchema == null || outputSchema.getFields() == null) {
endColumns = new ArrayList<>(0);
} else {
endColumns = new ArrayList<>(outputSchema.getFields().size());
for (Schema.Field field : outputSchema.getFields()) {
endColumns.add(field.getName());
}
}
FieldLevelLineage f;
store = new DefaultTransientStore();
registry = new CompositeDirectiveRegistry(
new SystemDirectiveRegistry(),
new UserDirectiveRegistry(context.getContext())
);
RecipeParser directives = new GrammarBasedParser(new MigrateToV2(config.directives).migrate(), registry);
ExecutorContext ctx = new WranglerPipelineContext(ExecutorContext.Environment.TRANSFORM,
context.getContext(), store);
try {
// Create the pipeline executor with context being set.
pipeline = new RecipePipelineExecutor();
pipeline.initialize(directives, ctx);
} catch (Exception e) {
throw new Exception(
String.format("Stage:%s - %s", context.getContext().getStageName(), e.getMessage())
);
}
try {
f = (new WranglerFieldLevelLineage.Builder(startColumns, endColumns)).build(pipeline.lineage());
} catch (LineageGenerationException e) {
LOG.warn(
String.format("Unable to generate lineage: %s", e.getMessage())
);
f = null;
}
LOG.info(
"Field-level lineage generated"
);
// context.recordLineage(f);
}
}
...
New REST APIs
| Path | Method | Description | Response |
|---|
/v3/namespaces/{namespace-id}/datasets/{dataset-id}/columns/{column-id}/lineage?start=<start-ts>&end=<end-ts>&maxLevels=<max-levels> | GET | Returns list of directives applied to the specified column in the specified dataset | 200: Successful Response TBD, but will contain a Tree representation |
/v3/namespaces/{namespace-id}/streams/{stream-id}/columns/{column-id}/lineage?start=<start-ts>&end=<end-ts>&maxLevels=<max-levels> | GET | Returns list of directives applied to the specified column in the specified stream | 200: Successful Response TBD, but will contain a Tree representation |
CLI Impact or Changes
TBD
UI Impact or Changes
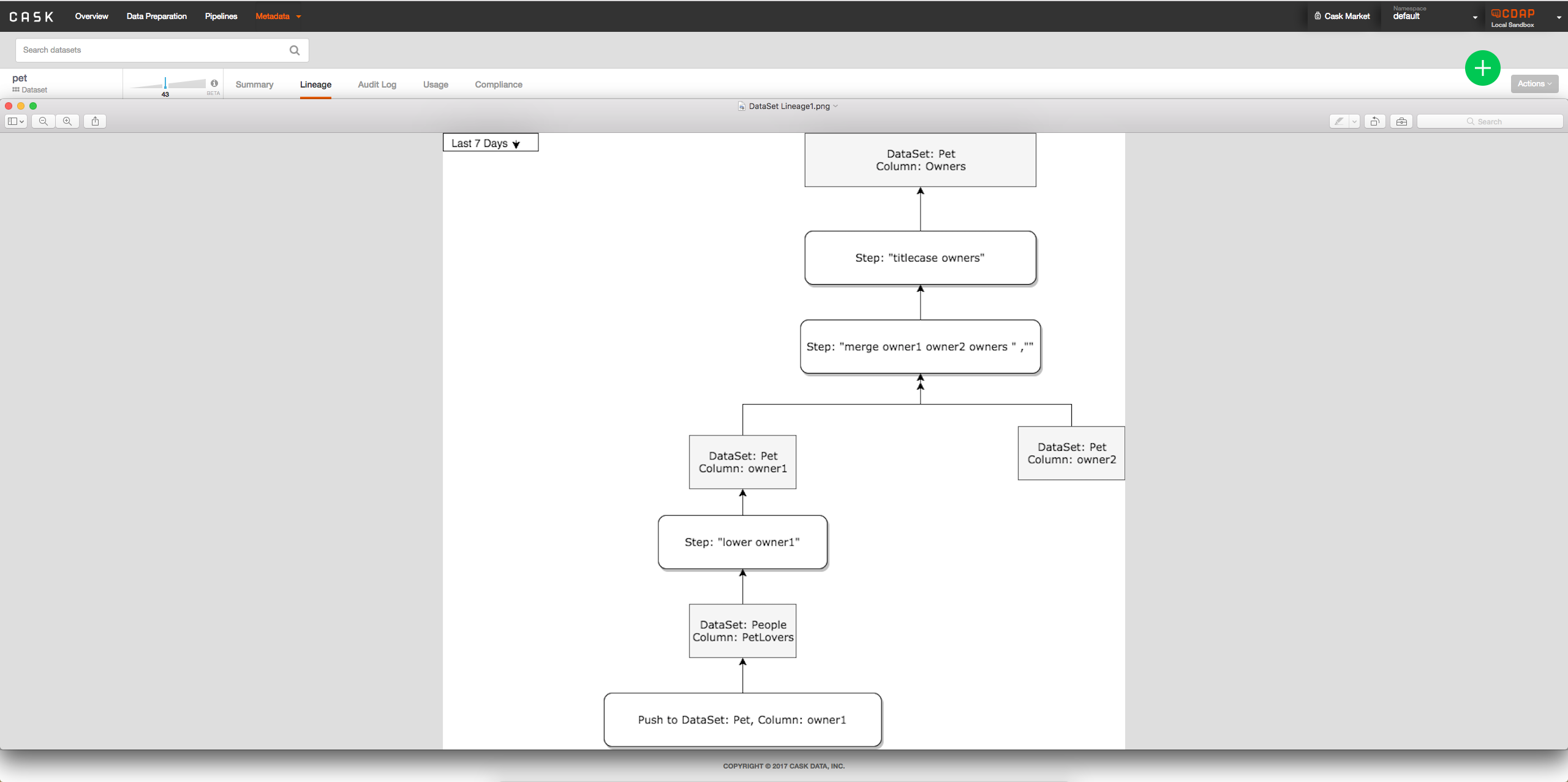
- Option 2: Add interface to metadata table when viewing dataset to see lineage of columns possibly by clicking on column: -> When a column is clicked on will look something like:
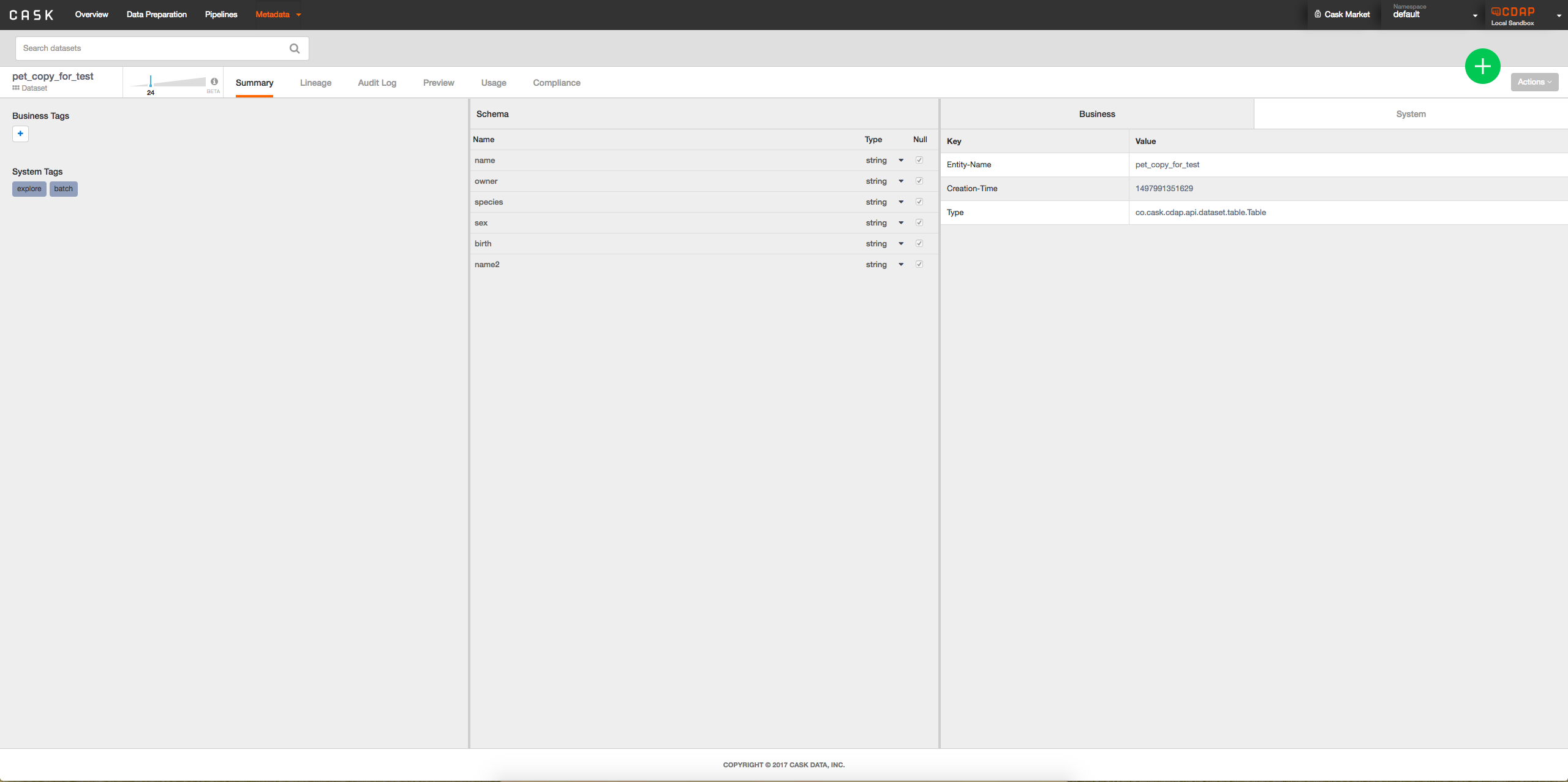 ->
-> 
- Option 2: Show all columns at once directly on lineage tab from clicking on dataset, tab between field level and dataset level:
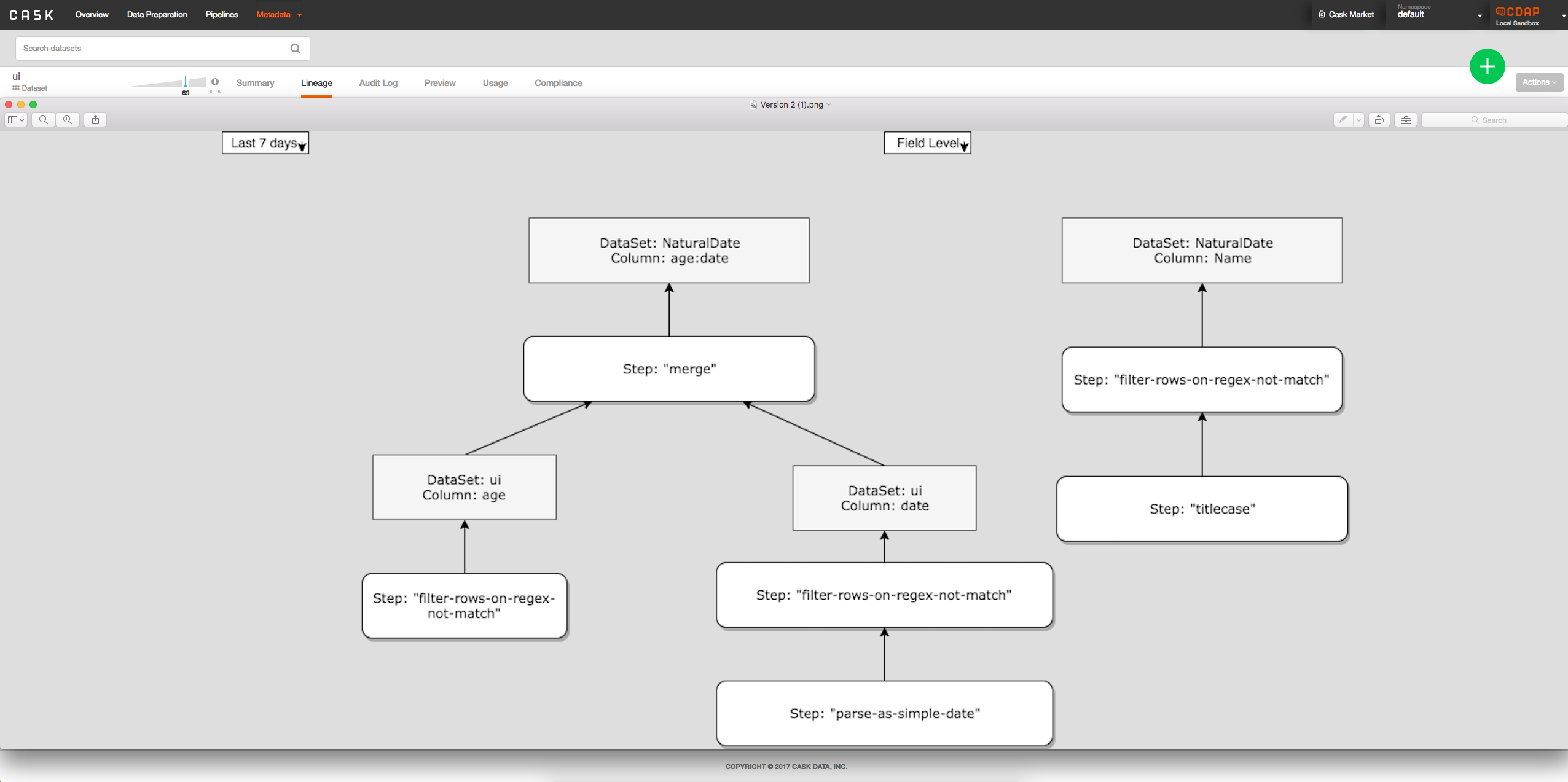
Security Impact
Should be none, TBD
Future Work
Taking this work to completion is seen in branches 'feature/field-level-lineage-api' and 'feature/field-level-lineage-graph'.
These branches show implementations of recordLineage as well as the graph that can be used to join many instances of FieldLevelLineage together.
Beginnings of designing data sets for storing the information in HBase is present in 'feature/field-level-lineage-graph'.
Impact on Infrastructure Outages
Storage in HBase. Key being dataset reference name + field name + ProgramRunId. For each pipeline a node is added to this key; Impact TBD.
Test Scenarios
| Test ID | Test Description | Expected Results |
|---|
| 1 | Tests all directives | All Directive subclasses should be properly parsed containing all correct columns with correct labels |
| 2 | Multiple datasets/streams | Lineages are correctly shown between different datasets/streams |
| 3 | Tests all build() | FieldLevelLineage.build() always correctly stores step |
Releases
Release 4.3.0
Release 4.4.0
- Fixing TextDirectives and parsing of directives in general






